library(MASS)
library(lmtest)
library(Ecdat)
library(sur)
library(plotly)
library(tidyverse)
library(gridExtra)Klausurvorbereitung
Libraries
Einfache Regressionsmodelle
Regressions with Boston Housing Data
housing <- Boston
?Boston
housing %>% ggplot(aes(x=lstat, y=medv))+
geom_point()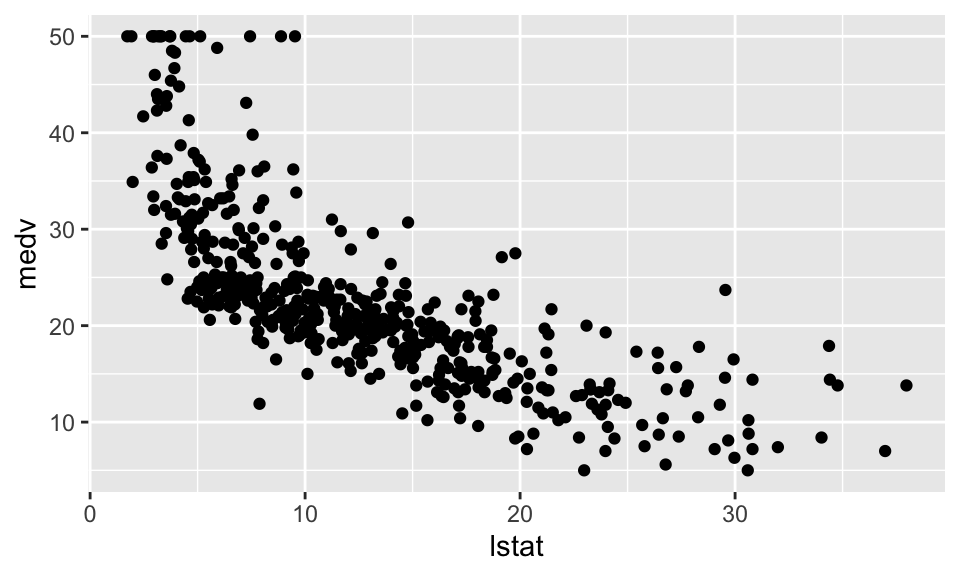
Regression Nummer 1 mit \(\text{medv} = \beta_1 + \beta_2 \cdot \text{lstat}\)
Reg1 <- lm(formula = medv ~ lstat+1, data = housing)
summary(Reg1)
Call:
lm(formula = medv ~ lstat + 1, data = housing)
Residuals:
Min 1Q Median 3Q Max
-15.168 -3.990 -1.318 2.034 24.500
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 34.55384 0.56263 61.41 <2e-16 ***
lstat -0.95005 0.03873 -24.53 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 6.216 on 504 degrees of freedom
Multiple R-squared: 0.5441, Adjusted R-squared: 0.5432
F-statistic: 601.6 on 1 and 504 DF, p-value: < 2.2e-16housing %>% ggplot(aes(x=lstat, y=medv))+
geom_point()+
geom_abline(intercept = Reg1$coefficients[1], slope = Reg1$coefficients[2])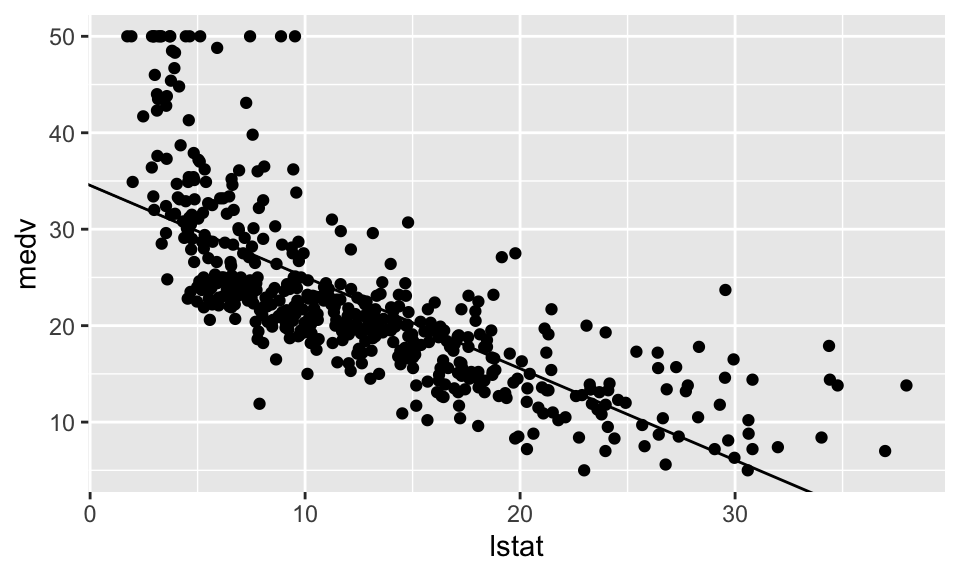
resettest(medv ~ lstat , power=2, type="regressor", data = housing)
RESET test
data: medv ~ lstat
RESET = 135.2, df1 = 1, df2 = 503, p-value < 2.2e-16Regression Nummer 2 mit \(\ln(\text{medv}) = \beta_1 + \beta_2 \cdot \text{lstat}\)
Reg2 <- lm(formula = log(medv)~lstat, data = housing)
summary(Reg2)
Call:
lm(formula = log(medv) ~ lstat, data = housing)
Residuals:
Min 1Q Median 3Q Max
-0.94921 -0.14838 -0.02043 0.11441 0.90958
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.617572 0.021971 164.65 <2e-16 ***
lstat -0.046080 0.001513 -30.46 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.2427 on 504 degrees of freedom
Multiple R-squared: 0.6481, Adjusted R-squared: 0.6474
F-statistic: 928.1 on 1 and 504 DF, p-value: < 2.2e-16housing %>% ggplot(aes(lstat, medv))+
geom_point()+
scale_y_continuous(trans = "log")+
geom_abline(intercept = Reg2$coefficients[1], slope = Reg2$coefficients[2])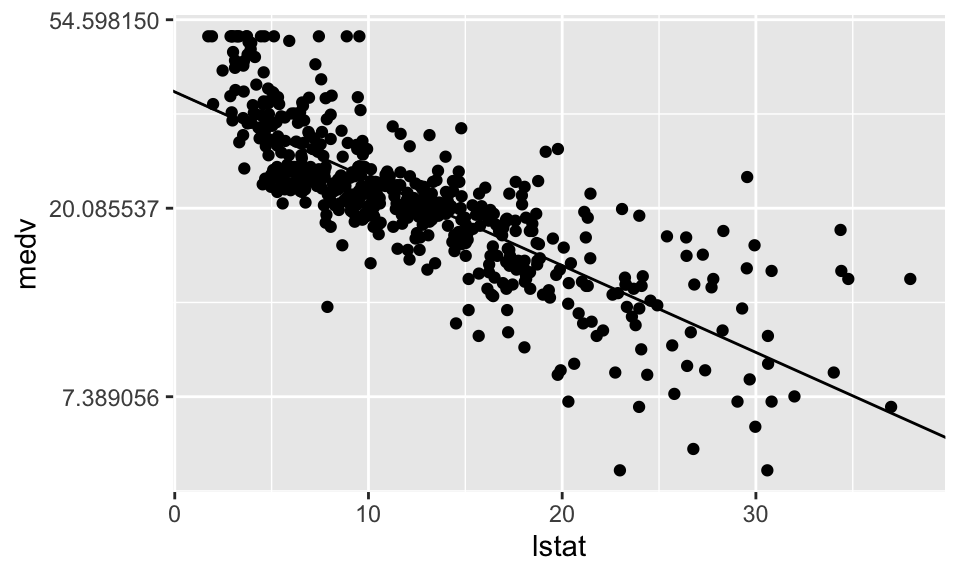
Regression Nummer 3 mit \(\ln(\text{medv}) = \beta_1 + \beta_2 \cdot \text{lstat} + \beta_3 \cdot \text{lstat}^2\)
Reg3 <- lm(formula = medv ~ lstat+I(lstat^2), data = housing)
summary(Reg3)
Call:
lm(formula = medv ~ lstat + I(lstat^2), data = housing)
Residuals:
Min 1Q Median 3Q Max
-15.2834 -3.8313 -0.5295 2.3095 25.4148
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 42.862007 0.872084 49.15 <2e-16 ***
lstat -2.332821 0.123803 -18.84 <2e-16 ***
I(lstat^2) 0.043547 0.003745 11.63 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 5.524 on 503 degrees of freedom
Multiple R-squared: 0.6407, Adjusted R-squared: 0.6393
F-statistic: 448.5 on 2 and 503 DF, p-value: < 2.2e-16housing %>% ggplot(aes(x=lstat, y=medv))+
geom_point()+
stat_function(fun = function(x)
{Reg3$coefficients[1] + Reg3$coefficients[2]*x + Reg3$coefficients[3]*x^2})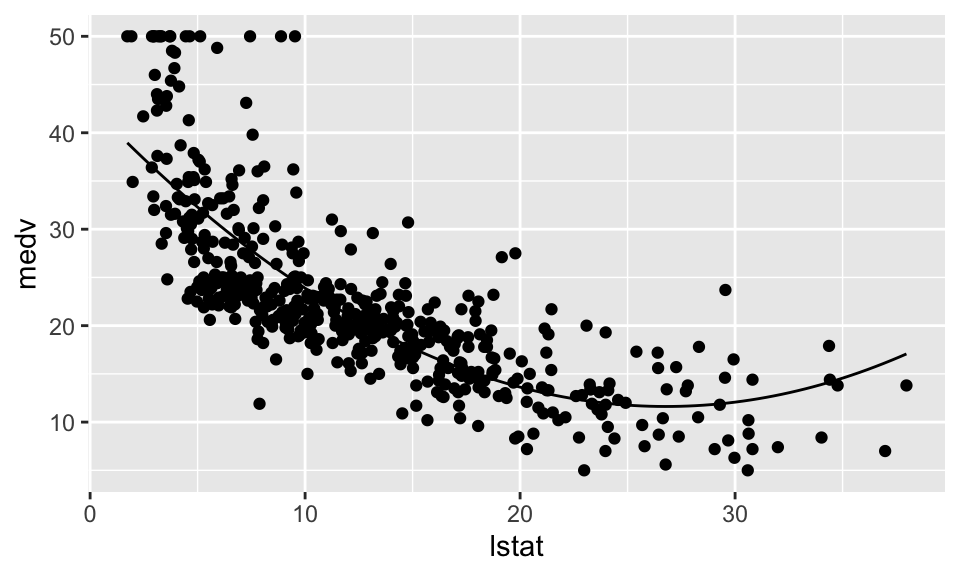
resettest(medv ~ lstat+I(lstat^2) , power=2, type="regressor", data = housing)
RESET test
data: medv ~ lstat + I(lstat^2)
RESET = 9.3452, df1 = 2, df2 = 501, p-value = 0.0001036Regression Nummer 4 mit \(\ln(\text{medv}) = \beta_1 + \beta_2 \cdot \text{lstat} + \beta_3 \cdot \text{lstat}^2 + \beta_4 \cdot \text{lstat}^3\)
Reg4 <- lm(formula = medv ~ lstat+I(lstat^2)+I(lstat^3), data = housing)
summary(Reg4)
Call:
lm(formula = medv ~ lstat + I(lstat^2) + I(lstat^3), data = housing)
Residuals:
Min 1Q Median 3Q Max
-14.5441 -3.7122 -0.5145 2.4846 26.4153
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 48.6496253 1.4347240 33.909 < 2e-16 ***
lstat -3.8655928 0.3287861 -11.757 < 2e-16 ***
I(lstat^2) 0.1487385 0.0212987 6.983 9.18e-12 ***
I(lstat^3) -0.0020039 0.0003997 -5.013 7.43e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 5.396 on 502 degrees of freedom
Multiple R-squared: 0.6578, Adjusted R-squared: 0.6558
F-statistic: 321.7 on 3 and 502 DF, p-value: < 2.2e-16housing %>% ggplot(aes(x=lstat, y=medv))+
geom_point()+
stat_function(fun = function(x)
{Reg4$coefficients[1] + Reg4$coefficients[2]*x + Reg4$coefficients[3]*x^2 + Reg4$coefficients[4]*x^3})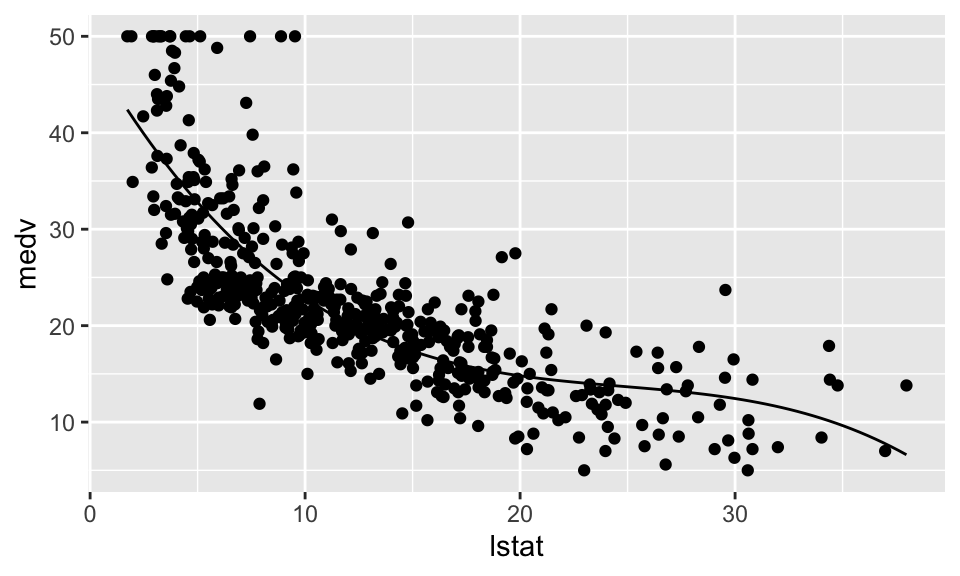
resettest(medv ~ lstat+I(lstat^2)+I(lstat^3) , power=2, type="regressor", data = housing)
RESET test
data: medv ~ lstat + I(lstat^2) + I(lstat^3)
RESET = 12.165, df1 = 3, df2 = 499, p-value = 1.076e-07Prüfen auf Einflussreiche Beobachtungen und Außreißer
length(housing$medv)[1] 506column1 <- seq(1,1, length.out=506)
matrix <- cbind(column1, housing$lstat)
hatMatrix <- matrix %*% solve(t(matrix) %*% matrix) %*% t(matrix)
diagHatMatrix <- diag(hatMatrix)
show(diagHatMatrix) [1] 0.004262518 0.002455527 0.004863680 0.005639779 0.004058706 0.004127513
[7] 0.001978217 0.003615365 0.013567169 0.002744185 0.004336932 0.001991064
[13] 0.002339159 0.002725692 0.002198662 0.002655757 0.003408468 0.002134252
[19] 0.002012300 0.002049494 0.004694701 0.002030073 0.003405580 0.004004395
[25] 0.002492748 0.002553939 0.002156943 0.002807608 0.001977123 0.001993876
[31] 0.005818324 0.001982098 0.010779804 0.003236561 0.004270793 0.002319519
[37] 0.002036287 0.002561791 0.002223479 0.004672735 0.006399736 0.004346707
[43] 0.003794663 0.003031568 0.002350192 0.002208052 0.002063299 0.003443523
[49] 0.014778002 0.002464813 0.002000947 0.002379670 0.004087240 0.002668814
[55] 0.002155272 0.004364946 0.003815983 0.004917504 0.003279450 0.002433949
[61] 0.001985874 0.002100279 0.003338594 0.002362339 0.002799052 0.004450983
[67] 0.002202395 0.002781274 0.001983698 0.002555776 0.003343198 0.002274893
[73] 0.003952049 0.002991470 0.003315691 0.002511647 0.001994402 0.002196808
[79] 0.001980090 0.002466502 0.004081517 0.003122517 0.003343198 0.003003418
[85] 0.002333513 0.003432148 0.001977947 0.002665538 0.003963144 0.003853592
[91] 0.002549791 0.002746302 0.002760198 0.003588295 0.002141560 0.003375643
[97] 0.002043235 0.004744393 0.005179954 0.003598318 0.002382178 0.002940504
[103] 0.002135213 0.002000332 0.001980337 0.002542020 0.003377449 0.002056463
[109] 0.001981983 0.002302167 0.001980959 0.002217636 0.002467572 0.002740735
[115] 0.002164752 0.002351126 0.001990879 0.002191290 0.002262928 0.002011844
[121] 0.002090755 0.002077809 0.003057586 0.008295685 0.002918905 0.002156943
[127] 0.010261454 0.002775582 0.002267164 0.003232141 0.001976394 0.001982284
[133] 0.002067549 0.002195675 0.002818424 0.002696596 0.002676666 0.002121969
[139] 0.004893136 0.003285699 0.007117935 0.020357684 0.009769828 0.009335944
[145] 0.012724338 0.010885361 0.002596636 0.013036672 0.011507565 0.004981295
[151] 0.002057583 0.001991547 0.001987319 0.002358400 0.002212604 0.002193833
[157] 0.002448425 0.004500830 0.003480090 0.003051908 0.003963144 0.006609388
[163] 0.006449610 0.005358735 0.002016137 0.002290159 0.005088907 0.001986506
[169] 0.002069946 0.002045290 0.002098895 0.001991359 0.002137400 0.002483198
[175] 0.002328817 0.004058706 0.002227414 0.003548513 0.003252596 0.004226903
[181] 0.002983544 0.002374680 0.004358858 0.003864408 0.002044657 0.001985874
[187] 0.004589259 0.003361691 0.004519651 0.004024722 0.004191568 0.004438598
[193] 0.005692773 0.004232820 0.004634045 0.005617183 0.004830292 0.002611037
[199] 0.003389664 0.004519651 0.004589259 0.003035620 0.005512663 0.005012891
[205] 0.005685179 0.002099742 0.002086282 0.003111519 0.002132690 0.006206176
[211] 0.002804019 0.006958334 0.002419106 0.002392283 0.013062901 0.002369720
[217] 0.002004800 0.002317214 0.003053492 0.002156295 0.002312627 0.004988131
[223] 0.002264222 0.002967784 0.004790484 0.004475844 0.005497855 0.003514111
[229] 0.004937816 0.005047327 0.002015354 0.004104453 0.006002898 0.004917504
[235] 0.002799052 0.002098361 0.002352606 0.004413923 0.003514111 0.003060098
[241] 0.002039218 0.001978771 0.002056031 0.004139089 0.001977194 0.003285699
[247] 0.002450086 0.002219576 0.002357457 0.003417917 0.003747147 0.005165862
[253] 0.005208233 0.005201152 0.003413189 0.002425985 0.005512663 0.004179852
[259] 0.002894623 0.003261516 0.002340615 0.003105701 0.003741906 0.002052728
[265] 0.002781274 0.002164752 0.002153608 0.003031568 0.005475703 0.002014878
[271] 0.001980959 0.003403756 0.002917424 0.003408468 0.005208233 0.005609667
[277] 0.003669351 0.004777276 0.003135210 0.004340643 0.005047327 0.004500830
[283] 0.005587165 0.005475703 0.002872102 0.002735962 0.001979263 0.003156521
[289] 0.002967784 0.002359894 0.005351491 0.005187012 0.004432418 0.002620491
[295] 0.002173404 0.003558412 0.003051908 0.002370678 0.004268481 0.004407774
[301] 0.003659110 0.002362339 0.002592337 0.004334587 0.003248147 0.002514534
[307] 0.003460820 0.002995445 0.004532237 0.002255825 0.001976291 0.003705438
[313] 0.002010091 0.002853548 0.002418091 0.002027913 0.003227728 0.002395817
[319] 0.002180465 0.001976514 0.003130971 0.003274955 0.002928929 0.002008658
[325] 0.003633644 0.004203315 0.003618458 0.001977013 0.002255825 0.003072442
[331] 0.002469266 0.001978217 0.002879578 0.003864408 0.003329410 0.002813413
[337] 0.002292371 0.002146402 0.002642825 0.002305805 0.002415475 0.003968703
[343] 0.002598539 0.003139457 0.004488322 0.002151313 0.001976296 0.003514111
[349] 0.003700260 0.003752395 0.003705438 0.003968703 0.002894623 0.004557503
[355] 0.002799052 0.003924448 0.002926574 0.001991064 0.002029720 0.001976296
[361] 0.002894623 0.002068011 0.002211862 0.002129588 0.004081517 0.003165100
[367] 0.002046734 0.001994079 0.005402365 0.005068082 0.005624707 0.002355028
[373] 0.002529089 0.020971010 0.026865167 0.002000332 0.006328634 0.004839537
[379] 0.006706492 0.005210978 0.002782645 0.004733829 0.006629662 0.007481612
[385] 0.014525439 0.014778002 0.011458958 0.016496011 0.014511481 0.004585358
[391] 0.002747643 0.003424489 0.008566016 0.002222280 0.002507007 0.002751108
[397] 0.003728250 0.004026908 0.014469655 0.013620902 0.009714913 0.004258869
[403] 0.004252918 0.003943126 0.010398143 0.006117484 0.006411243 0.001986909
[409] 0.009314576 0.003948657 0.002227414 0.004826215 0.020290158 0.004118201
[415] 0.024956703 0.012416477 0.008677773 0.009573043 0.004440995 0.005927236
[421] 0.002193833 0.002336788 0.002057583 0.006369841 0.002765047 0.007325531
[427] 0.002334426 0.002111630 0.005029309 0.007046690 0.002942004 0.003899157
[433] 0.001991359 0.002470338 0.002222280 0.006353335 0.003107324 0.009368054
[439] 0.019704605 0.006037670 0.005449119 0.003807372 0.002578151 0.003467489
[445] 0.006792596 0.006958334 0.003000972 0.002533161 0.003141104 0.003697091
[451] 0.002866097 0.002977175 0.002804019 0.002624888 0.003400872 0.003141104
[457] 0.003545487 0.002689921 0.002473112 0.002138986 0.002527295 0.002131135
[463] 0.002045692 0.002193122 0.001988766 0.002060989 0.002761550 0.004893136
[469] 0.003141104 0.002148664 0.002489920 0.001978112 0.002089425 0.002014579
[475] 0.003145361 0.007064455 0.003386795 0.007810023 0.003098957 0.001984392
[481] 0.002118400 0.002913604 0.003212838 0.002169920 0.001994608 0.002143166
[487] 0.002186542 0.002032488 0.003111519 0.006949541 0.013234152 0.003115722
[493] 0.001995146 0.001992343 0.002010373 0.002926574 0.004773237 0.002057583
[499] 0.001979052 0.002208788 0.002085483 0.002321832 0.002472037 0.003886132
[505] 0.003456022 0.002860947# jetzt müsste geprüft werden welche Zeilen der diagHatMatrix größer sind als 2*(2/506)
as.data.frame(diagHatMatrix) %>%
mutate(n = 1:nrow(.)) %>%
filter(diagHatMatrix > 2*(2/506))Auf Koliniarität prüfen
Reg5 <- lm(formula = log(medv) ~ lstat + crim, data = housing)
summary(Reg5)
Call:
lm(formula = log(medv) ~ lstat + crim, data = housing)
Residuals:
Min 1Q Median 3Q Max
-0.78707 -0.15045 -0.02918 0.11439 0.88271
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.585383 0.021417 167.409 < 2e-16 ***
lstat -0.040776 0.001620 -25.175 < 2e-16 ***
crim -0.009665 0.001345 -7.187 2.4e-12 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.2314 on 503 degrees of freedom
Multiple R-squared: 0.6809, Adjusted R-squared: 0.6796
F-statistic: 536.5 on 2 and 503 DF, p-value: < 2.2e-16plot_ly(x=housing$lstat, y=housing$crim, z=log(housing$medv),
type="scatter3d", mode="markers", color=housing$lstat, size=15)Ein paar Tests
Einfache Regression
n <- 40
quadratmeter <- seq(n)
preis <- rnorm(n,0+3*quadratmeter, 40)
data <- data.frame(quadratmeter, preis)
a1 <- lm(preis~quadratmeter)
summary(a1)
Call:
lm(formula = preis ~ quadratmeter)
Residuals:
Min 1Q Median 3Q Max
-82.762 -34.350 3.379 32.215 70.983
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 24.5103 13.1134 1.869 0.06933 .
quadratmeter 1.8730 0.5574 3.360 0.00178 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 40.69 on 38 degrees of freedom
Multiple R-squared: 0.2291, Adjusted R-squared: 0.2088
F-statistic: 11.29 on 1 and 38 DF, p-value: 0.001783data %>% ggplot(aes(quadratmeter, preis))+
geom_point()+
stat_function(fun = function(x) {a1$coefficients[1] + a1$coefficients[2]*x})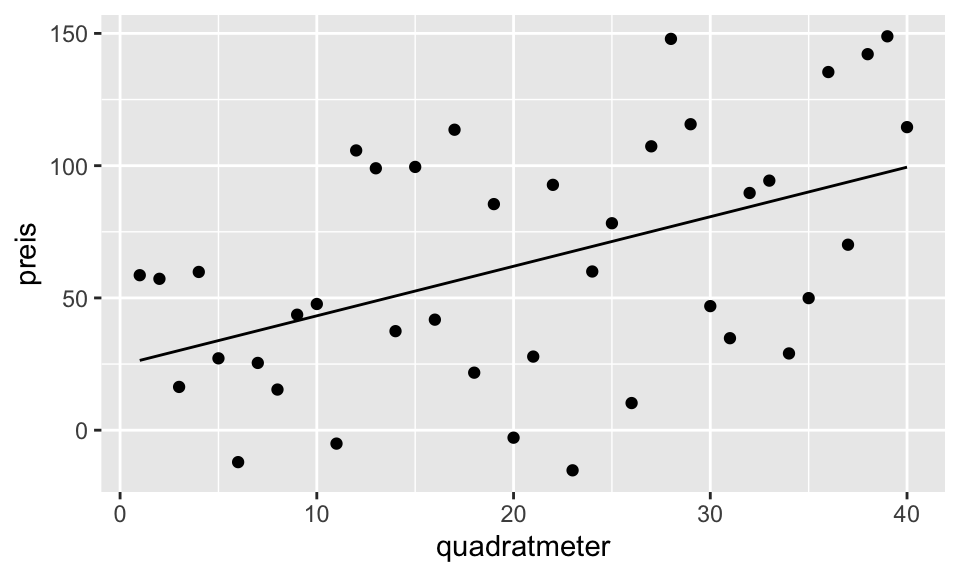
Heteroskedastizität
Generierung von Heteroskedastizität und Verteilung der Residuen
n2 <- 10000
x <- runif(n2, min = 0, max = 10)
y <- rnorm(n2, mean = 50+3*x, sd = 5+2*x)
df1 <- data.frame(x, y)
df1 %>%
ggplot(aes(x, y))+
geom_point(alpha = .5, size = .5)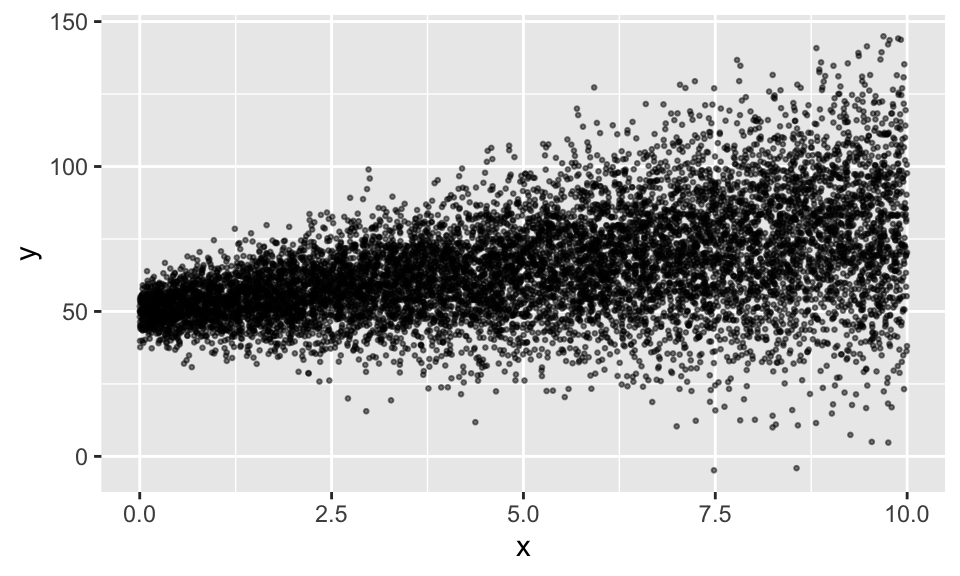
a2 <- lm(y~x)
summary(a2)
Call:
lm(formula = y ~ x)
Residuals:
Min 1Q Median 3Q Max
-79.653 -9.003 0.068 8.915 65.926
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 49.91845 0.32474 153.72 <2e-16 ***
x 2.99971 0.05626 53.32 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 16.15 on 9998 degrees of freedom
Multiple R-squared: 0.2214, Adjusted R-squared: 0.2213
F-statistic: 2843 on 1 and 9998 DF, p-value: < 2.2e-16a2 %>% ggplot(aes(x=a2$residuals))+
geom_density()+
stat_function(fun = function(x){dnorm(x, mean = mean(a2$residuals), sd = sd(a2$residuals))}, linetype = "dashed")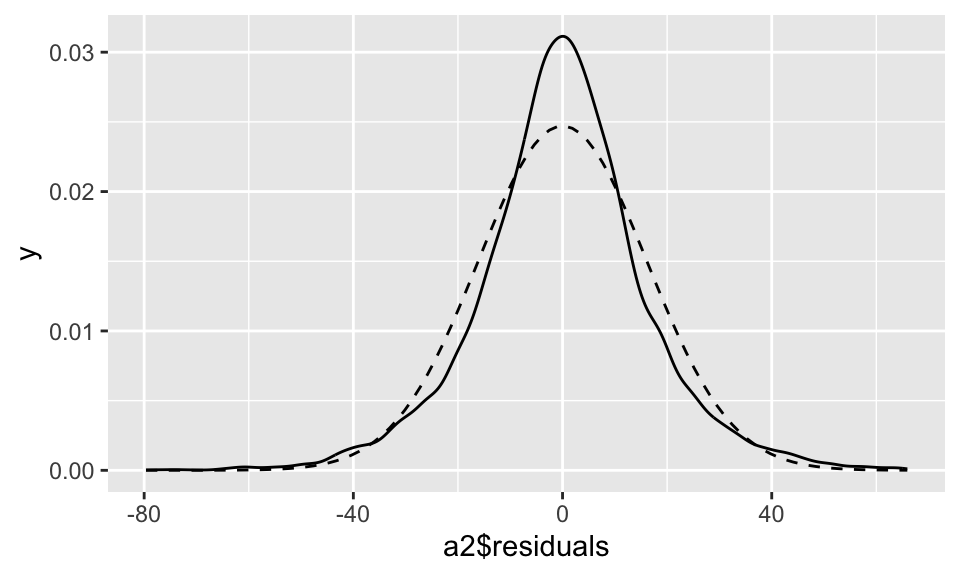
Die Residuen (durchgezogene Linie) sind nicht normalverteilt (gestrichelte Linie)
Beispiel einer fehlehnden Binärvariable
n <- 500
data <- data.frame(x = runif(n, min = 0, max = 10),
group = c(1,2)) %>%
mutate(group = as.factor(group),
y = if_else(group == 1,
5 + 3*x + rnorm(n, 0, 2),
20 + 3*x + rnorm(n, 0, 2)))
data %>%
ggplot(aes(x = x, y = y, color = group))+
geom_point()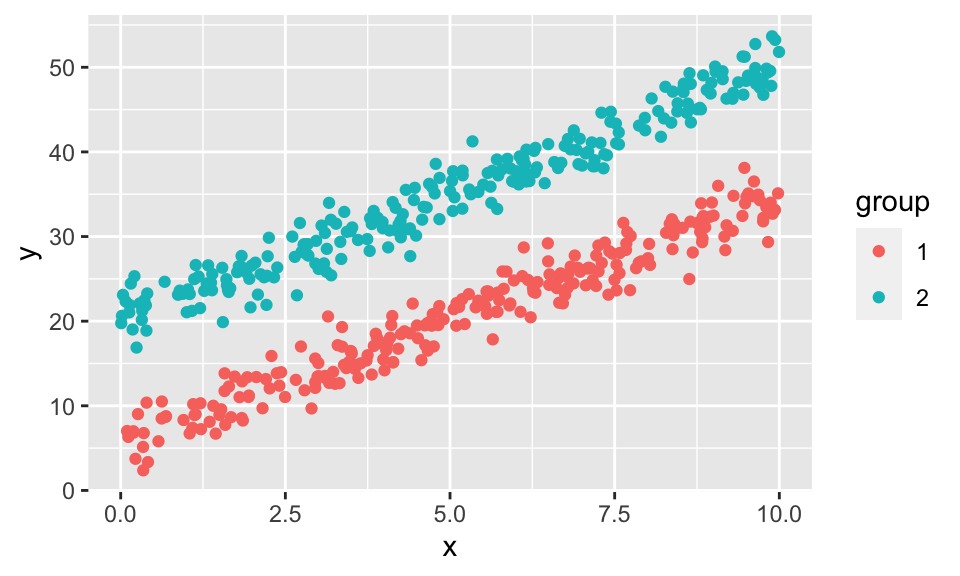
reg1 <- lm(y ~ x, data = data)
reg1
Call:
lm(formula = y ~ x, data = data)
Coefficients:
(Intercept) x
13.315 2.846 data.frame(resid = resid(reg1)) %>%
ggplot(aes(x = resid))+
geom_histogram(bins = 30)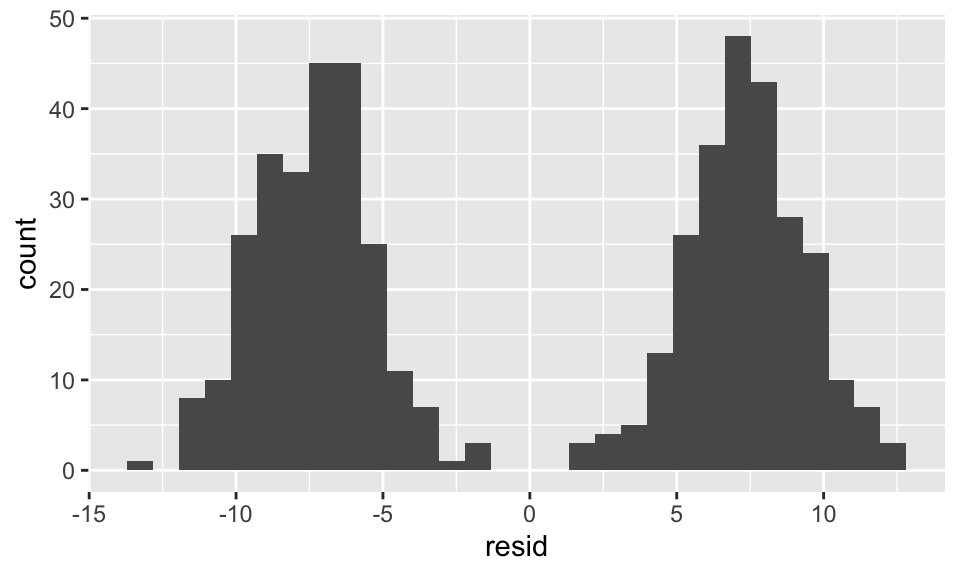
reg2 <- lm(y ~ x + group, data = data)
reg2
Call:
lm(formula = y ~ x + group, data = data)
Coefficients:
(Intercept) x group2
5.326 2.957 14.839 data.frame(resid = resid(reg2)) %>%
ggplot(aes(x = resid))+
geom_histogram(bins = 30)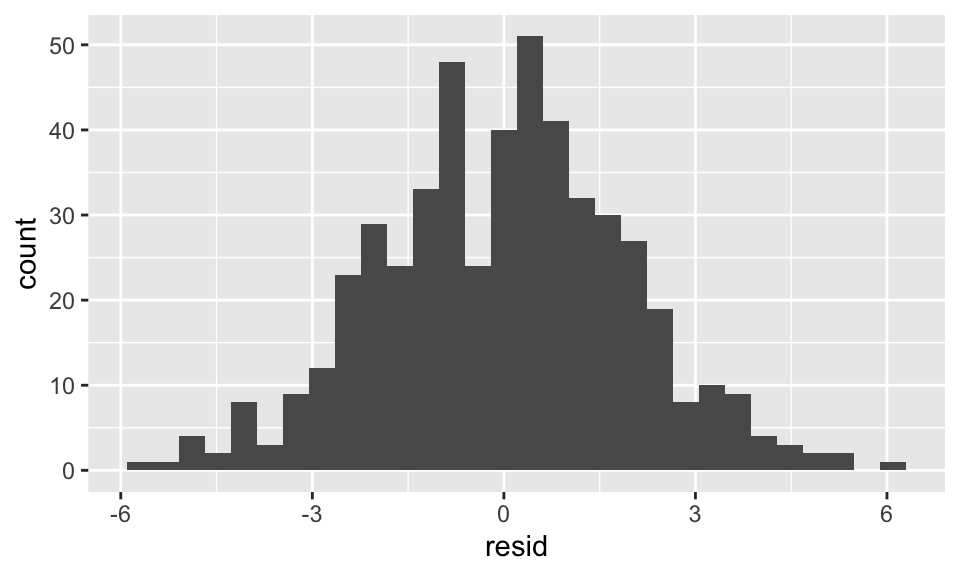
Chow Test für Strukturbruch
Wir generieren zuerst einen Datensatz mit einem Strukturbruch bei \(x=0\). Es soll nicht zu offensichtlich sein, damit der F-Test am Ende noch interessant bleibt.
n <- 200
x1 <- runif(n/2, min = -5, max = 0)
x2 <- runif(n/2, min = 0, max = 5)
y1 <- 5 + 1*x1 + rnorm(n/2, 0, 3)
y2 <- 5 + 3*x2 + rnorm(n/2, 0, 3)
data <- data.frame(x = c(x1, x2), y = c(y1, y2))
data %>%
ggplot(aes(x, y))+
geom_point()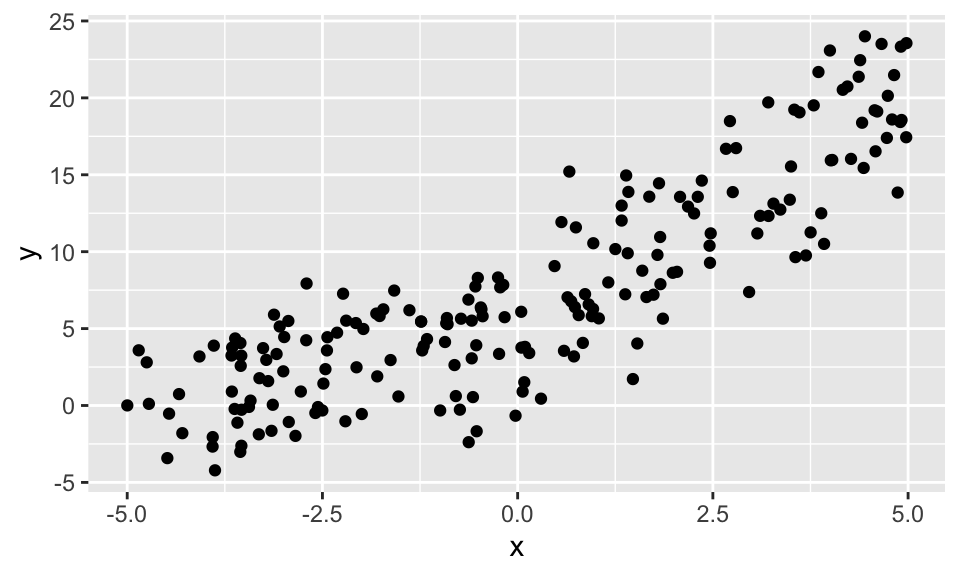
Wir sehen, dass eine “naive” lineare Regression hier kein gutes Ergebnis bringt.
reg1 <- lm(y ~ x, data = data)
summary(reg1)
Call:
lm(formula = y ~ x, data = data)
Residuals:
Min 1Q Median 3Q Max
-8.6773 -2.4415 -0.0878 2.4663 7.4319
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 7.32257 0.24542 29.84 <2e-16 ***
x 2.08198 0.08697 23.94 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.468 on 198 degrees of freedom
Multiple R-squared: 0.7432, Adjusted R-squared: 0.7419
F-statistic: 573.1 on 1 and 198 DF, p-value: < 2.2e-16data %>% ggplot(aes(x, y))+
geom_point()+
geom_abline(intercept = reg1$coefficients[1], slope = reg1$coefficients[2])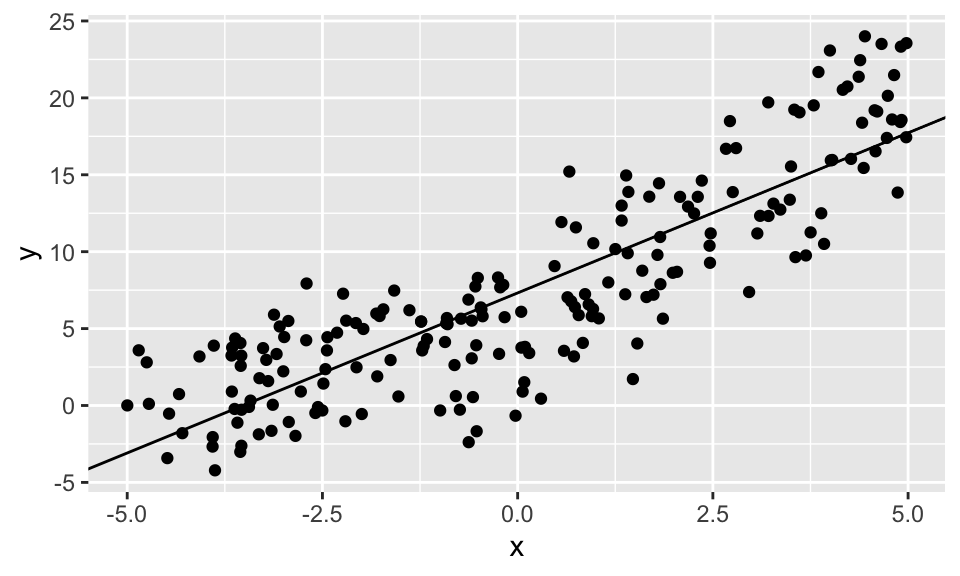
reg2.1 <- lm(y ~ x, data = data %>% filter(x < 0))
reg2.2 <- lm(y ~ x, data = data %>% filter(x > 0))
data %>% ggplot(aes(x, y))+
geom_point()+
geom_abline(intercept = reg2.1$coefficients[1], slope = reg2.1$coefficients[2])+
geom_abline(intercept = reg2.2$coefficients[1], slope = reg2.2$coefficients[2])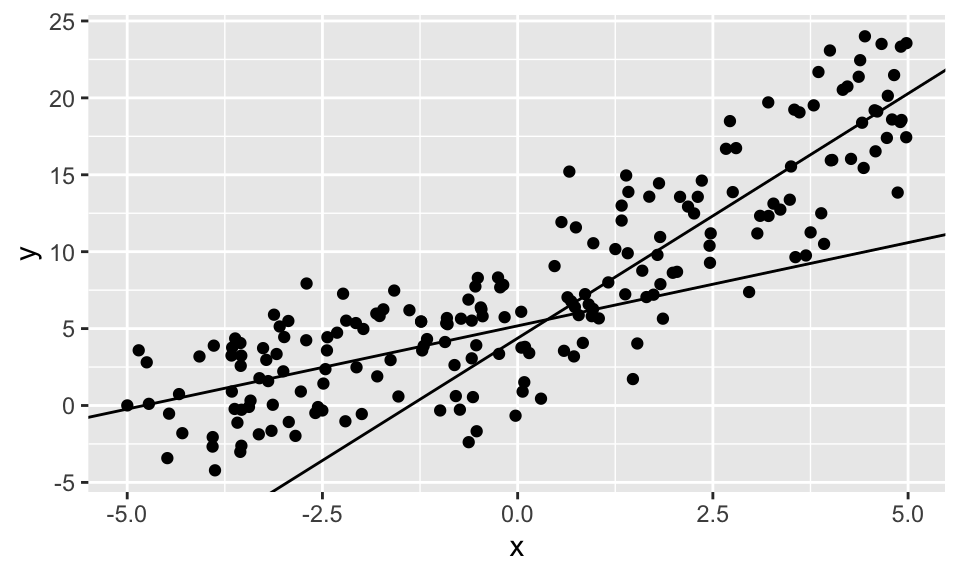
Der Chow Test zeigt ganz klar, dass ein Strukturbruch vorliegt.
SSR.R <- sum(resid(reg1)^2)
SSR.U <- sum(resid(reg2.1)^2) + sum(resid(reg2.2)^2)
Fstatistic <- ((SSR.R-SSR.U)/reg1$rank)/(SSR.U/(n - 2*reg1$rank))
Fstatistic[1] 24.326381-pf(Fstatistic, reg1$rank, n - 2*reg1$rank)[1] 3.657481e-10Echte Daten
Aus den Übungen
data("Bwages")
d <- Bwages
?Bwages
#d
data("Caschool")
#Caschool
?Caschool
data("Icecream")
#Icecream
?IcecreamReg1 <- lm(formula = cons~price+temp+I(price*temp), data = Icecream)
summary(Reg1)
Call:
lm(formula = cons ~ price + temp + I(price * temp), data = Icecream)
Residuals:
Min 1Q Median 3Q Max
-0.093826 -0.021935 0.006617 0.020962 0.082094
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.17029 0.89149 -0.191 0.850
price 1.38220 3.23280 0.428 0.672
temp 0.01880 0.01755 1.071 0.294
I(price * temp) -0.05731 0.06375 -0.899 0.377
Residual standard error: 0.04146 on 26 degrees of freedom
Multiple R-squared: 0.6439, Adjusted R-squared: 0.6028
F-statistic: 15.67 on 3 and 26 DF, p-value: 5.07e-06plot(Icecream$price ,Icecream$cons)
Reg2 <- lm(formula = cons~temp, data = Icecream)
summary(Reg2)
Call:
lm(formula = cons ~ temp, data = Icecream)
Residuals:
Min 1Q Median 3Q Max
-0.069411 -0.024478 -0.007371 0.029126 0.120516
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.2068621 0.0247002 8.375 4.13e-09 ***
temp 0.0031074 0.0004779 6.502 4.79e-07 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.04226 on 28 degrees of freedom
Multiple R-squared: 0.6016, Adjusted R-squared: 0.5874
F-statistic: 42.28 on 1 and 28 DF, p-value: 4.789e-07Reg2.1 <- lm(testscr~I(expnstu/1000)+compstu, data = Caschool)
summary(Reg2.1)
Call:
lm(formula = testscr ~ I(expnstu/1000) + compstu, data = Caschool)
Residuals:
Min 1Q Median 3Q Max
-52.815 -13.787 -0.116 13.078 47.016
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 625.000 7.528 83.024 < 2e-16 ***
I(expnstu/1000) 3.723 1.468 2.537 0.0116 *
compstu 68.993 14.323 4.817 2.04e-06 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 18.25 on 417 degrees of freedom
Multiple R-squared: 0.08736, Adjusted R-squared: 0.08299
F-statistic: 19.96 on 2 and 417 DF, p-value: 5.272e-09Reg2.2 <- lm(testscr~I(expnstu/1000), data = Caschool)
summary(Reg2.2)
Call:
lm(formula = testscr ~ I(expnstu/1000), data = Caschool)
Residuals:
Min 1Q Median 3Q Max
-50.146 -14.206 0.689 13.513 50.127
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 623.617 7.720 80.783 < 2e-16 ***
I(expnstu/1000) 5.749 1.443 3.984 7.99e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 18.72 on 418 degrees of freedom
Multiple R-squared: 0.03659, Adjusted R-squared: 0.03428
F-statistic: 15.87 on 1 and 418 DF, p-value: 7.989e-05Sterbedaten Deutschland
Quelle: Genesis Tabelle 12613-0003
data <- read_csv2("/Users/sebastiangeis/Downloads/12613-0003_$F_flat (1).csv") %>%
janitor::clean_names() %>%
select(jahr = zeit, geschlecht = x2_auspraegung_code, alter = x3_auspraegung_code, anzahl = bev002_gestorbene_anzahl) %>%
mutate(geschlecht = str_extract(geschlecht, ".{1}$") %>% factor(),
alter = str_extract(alter, "\\d{3}$") %>% as.numeric(),
anzahl = as.numeric(anzahl)) %>%
drop_na()
head(data)data_uncounted <- data %>%
uncount(anzahl)
head(data_uncounted)Der originale Datensatz gibt die Anzahl an Toten je Jahr, Alter und Geschlecht an. Wir transformieren den Datensatz, damit jede Zeile einer Person entspricht. Diese Datensatz hat dann 60356559 Zeilen. Das ist sehr viel, daher verwenden wir für die Regressionen jeweils nur eine Stichprobe.
data_uncounted %>%
group_by(jahr, geschlecht) %>%
summarise(alter = mean(alter)) %>%
ggplot(aes(x = jahr, y = alter, color = geschlecht))+
geom_line()`summarise()` has grouped output by 'jahr'. You can override using the
`.groups` argument.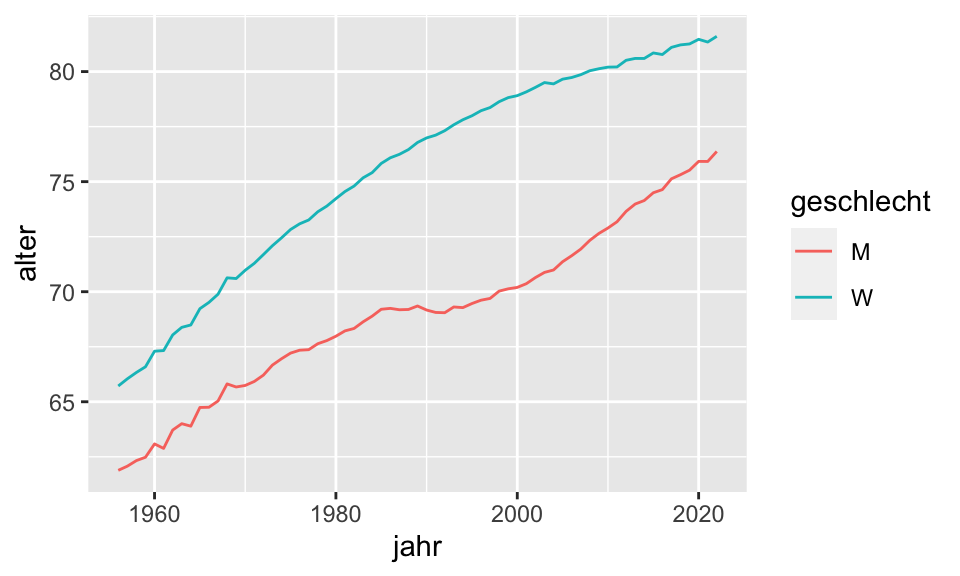
reg1 <- lm(alter ~ jahr, data = data_uncounted %>% slice_sample(n = 100000))
summary(reg1)
Call:
lm(formula = alter ~ jahr, data = data_uncounted %>% slice_sample(n = 1e+05))
Residuals:
Min 1Q Median 3Q Max
-79.582 -6.217 3.708 10.867 32.746
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -3.551e+02 5.380e+00 -66.00 <2e-16 ***
jahr 2.150e-01 2.705e-03 79.48 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 16.56 on 99998 degrees of freedom
Multiple R-squared: 0.05941, Adjusted R-squared: 0.0594
F-statistic: 6317 on 1 and 99998 DF, p-value: < 2.2e-16reg2 <- lm(alter ~ jahr + geschlecht, data = data_uncounted %>% slice_sample(n = 100000))
summary(reg2)
Call:
lm(formula = alter ~ jahr + geschlecht, data = data_uncounted %>%
slice_sample(n = 1e+05))
Residuals:
Min 1Q Median 3Q Max
-82.604 -5.804 3.553 10.368 34.847
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -3.531e+02 5.293e+00 -66.71 <2e-16 ***
jahr 2.123e-01 2.661e-03 79.78 <2e-16 ***
geschlechtW 6.439e+00 1.031e-01 62.48 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 16.29 on 99997 degrees of freedom
Multiple R-squared: 0.09372, Adjusted R-squared: 0.0937
F-statistic: 5170 on 2 and 99997 DF, p-value: < 2.2e-16data.frame(reg1 = reg1$residuals, reg2 = reg2$residuals) %>%
pivot_longer(cols = everything(), names_to = "model", values_to = "residuals") %>%
ggplot(aes(x = residuals, color = model))+
geom_density()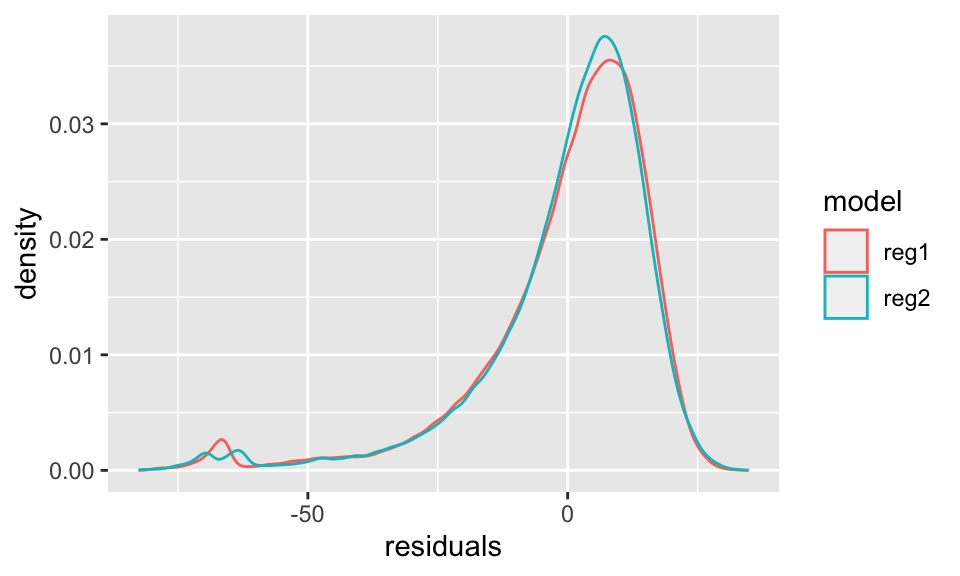
Betrachten wir den Plot mit dem durchschnittlichen Sterbealter, wird sofort klar, dass eine Binärvariable für das Geschlecht genutzt werden muss. In der Regression sehen wir dann, dass Frauen im Schnitt 6,6 Jahre länger leben als Männer.
Allerdings ist der Unterschied im Residuenplot nicht eindeutig. Für Regression 1, also ohne Berücksichtigung des Geschlechts, hätten wir zwei “Gipfel” erwartet. Dies ist nicht der Fall. Außerdem müssen wir die Verteilungsannahme überdenken, da die Residuen offensichtlich nicht normalverteilt sint. Hieran ändert auch eine log-Transofrmation nichts (nicht dargestellt).